Use the following button if you want to resubmit the job with
altered input or parameterization:
Input and runtime details for job 6107713 (precomputed example)
Sequence and Structure
 Sequence Input in FASTA Format Sequence Input in FASTA Format | [.fa] |
Alignment
 Alignment Type Alignment Type | Global | ||
 Alignment Mode Alignment Mode | no |
Alignment Scoring
 Structure Weight Structure Weight | 200 | ||
 Indel Opening Score Indel Opening Score | -800 | ||
 Indel Score Indel Score | -50 | ||
 Use RIBOSUM Use RIBOSUM | yes | ||
 Match Score Match Score | 50 | ||
 Mismatch Score Mismatch Score | 0 |
RNA Folding
Heuristics for speed/accuracy tradeoff
 Minimal Pair Probability Minimal Pair Probability | 5.0E-4 | ||
 Minimal Probability for Guide Tree Construction Minimal Probability for Guide Tree Construction | 0.005 | ||
 Maximal Difference for Sizes of Matched Arcs Maximal Difference for Sizes of Matched Arcs | 30 | ||
 Maximal Difference for Alignment Edges Maximal Difference for Alignment Edges | 60 |
Other Parameters
 Ignore Constraints Ignore Constraints | no | ||
 Disallow Lonely Pairs Disallow Lonely Pairs | yes | ||
 Use alifold consensus dot plots Use alifold consensus dot plots | yes | ||
 Consistency Transformation (LocARNA-P only) Consistency Transformation (LocARNA-P only) | no | ||
 Keep Sequence Input Order Keep Sequence Input Order | no | ||
 Stockholm alignment output Stockholm alignment output | yes |
Job ID 6107713 (server version 4.4.4)
 | Job Submitted & Queued | @ Thu Jun 08 13:26:13 CEST 2017 | |
 | LocARNA Started | @ Thu Jun 08 13:26:18 CEST 2017 | |
 | LocARNA Finished & Post-Processing | @ Thu Jun 08 13:26:21 CEST 2017 | |
 | Post-Processing Finished | @ Thu Jun 08 13:26:26 CEST 2017 | |
 | Job Completed | @ Thu Jun 08 13:26:26 CEST 2017 |
DIRECT ACCESS: http://rna.informatik.uni-freiburg.de/RetrieveResults.jsp?jobID=6107713&toolName=LocARNA ( 30 days expiry )
Description of the job
tRNA alignment
Output download complete results [zip]
 Alignment annotated with RNAalifold consensus structure
Alignment annotated with RNAalifold consensus structure
download alignment
[ClustalW]
[Stockholm]
[FASTA]
[structure]
[structure-pairs]
download alignment figure
[.pdf]
[.ps]
[.png]
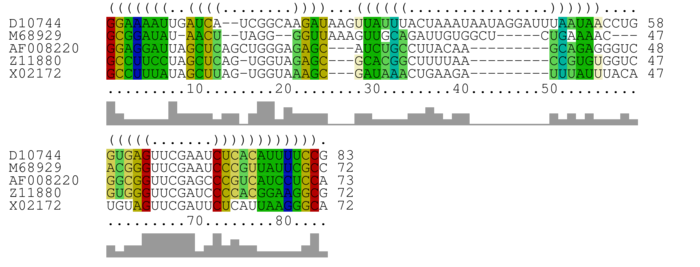
Bases are encoded by IUPAC codes. For interpretation of the colors see this legend. For a detailed description of the output, please see the help page.
 Guide tree and Intermediate Alignments
Guide tree and Intermediate Alignments
download guide tree
[NEWICK format]
[.svg]
download
[similarity matrix]
Intermediate Alignments
- intermediates/intermediate1.aln = D10744 M68929
- intermediates/intermediate2.aln = AF008220 Z11880
- intermediates/intermediate3.aln = AF008220 Z11880 X02172
- intermediates/intermediate4.aln = D10744 M68929 AF008220 Z11880 X02172
 RNAalifold consensus structure
RNAalifold consensus structure
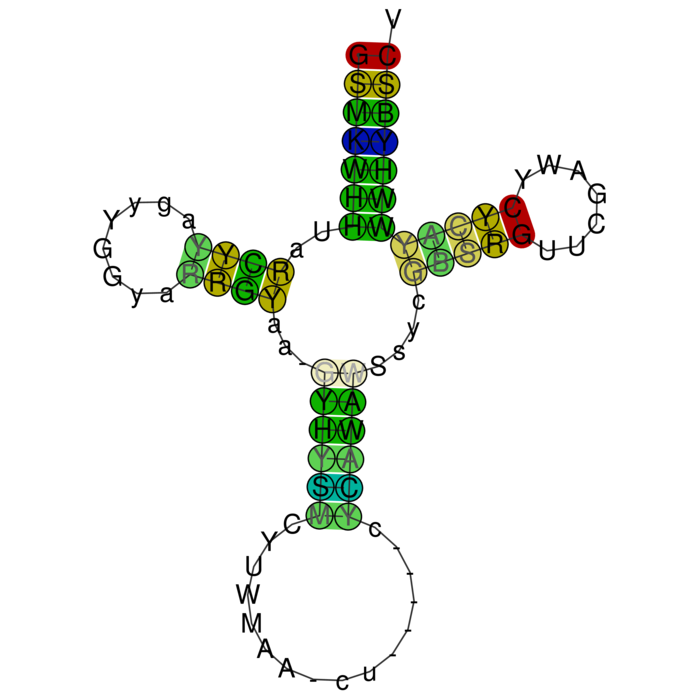
For interpretation of the colors see this legend. For a detailed description of the output, please see the help page.
Color Legend
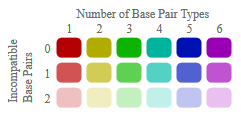
Compatible base pairs are colored, where the hue shows the number of different types C-G, G-C, A-U, U-A, G-U or U-G of compatible base pairs in
the corresponding columns. In this way the hue shows sequence conservation of the base pair. The saturation decreases with the number of
incompatible base pairs. Thus, it indicates the structural conservation of the base pair.
Job resubmission
 usability assessment
usability assessment
When using LocARNA please cite :
- Sebastian Will, Tejal Joshi, Ivo L. Hofacker, Peter F. Stadler, and Rolf Backofen.
LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs
RNA, 18 no. 5, pp. 900-14, 2012 - Sebastian Will, Kristin Reiche, Ivo L. Hofacker, Peter F. Stadler, and Rolf Backofen.
Inferring non-coding RNA families and classes by means of genome-scale structure-based clustering
PLoS Computational Biology, 3 no. 4, pp. e65, 2007 - Martin Raden, Syed M Ali, Omer S Alkhnbashi, Anke Busch, Fabrizio Costa, Jason A Davis, Florian Eggenhofer, Rick Gelhausen, Jens Georg, Steffen Heyne, Michael Hiller, Kousik Kundu, Robert Kleinkauf, Steffen C Lott, Mostafa M Mohamed, Alexander Mattheis, Milad Miladi, Andreas S Richter, Sebastian Will, Joachim Wolff, Patrick R Wright, and Rolf Backofen
Freiburg RNA tools: a central online resource for RNA-focused research and teaching
Nucleic Acids Research, 46(W1), W25-W29, 2018.
Results are computed with LocARNA version 1.9.1 linking Vienna RNA package 2.3.2



