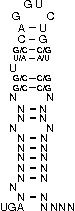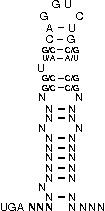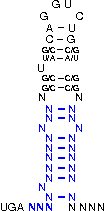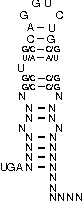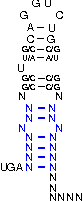Introduction
SECISDesign is a server for the design of SECIS-elements within the coding sequence of an
mRNA with both structure and sequence constraints.
This will trigger the insertion of a selenocystein at a preceding STOP codon.
Furthermore, a certain similarity to the original protein sequence is kept.
It can be used e.g. for recombinant expression of selenoproteins in E. coli.
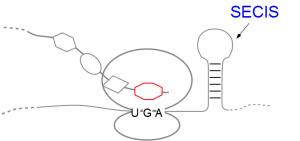
A SECIS-element (SEC Insertion Sequence) is an mRNA motif with both structural and sequential constraints,
that is required for the insertion of selenocysteine into a protein. Selenocysteine (Sec) is the rare 21st amino acid
and is incorporated in a particular class of proteins, called selenoproteins. Selenocysteine is encoded by the
UGA-codon, which is usually a STOP-codon. It has been shown that, in the case of selenocysteine, termination of
translation is inhibited in the presence of a specific mRNA sequence in the 3'-region after the UGA-codon that
forms a hairpin-like structure (the SECIS-element).
Selenoproteins have gained much interest, since they are of fundamental importance to human health and an
essential component of several major metabolic pathways, such as antioxidant defence systems, the thyroid
hormone metabolism, and the immune function. For this reason, there is an enormous interest in the catalytic
properties of selenoproteins, especially since a selenoprotein has greatly enhanced enzymatic activity compared
to its cysteine homologue.
Note: SECISDesign is not maintained anymore.
When using SECISDesign please cite :
- Anke Busch, Sebastian Will, and Rolf Backofen
SECISDesign - A Server to Design SECIS-Elements within the Coding Sequence
Bioinformatics, 2005, 21(15), 3312-3.
- Martin Raden, Syed M Ali, Omer S Alkhnbashi, Anke Busch, Fabrizio Costa, Jason A Davis, Florian Eggenhofer, Rick Gelhausen, Jens Georg, Steffen Heyne, Michael Hiller, Kousik Kundu, Robert Kleinkauf, Steffen C Lott, Mostafa M Mohamed, Alexander Mattheis, Milad Miladi, Andreas S Richter, Sebastian Will, Joachim Wolff, Patrick R Wright, and Rolf Backofen
Freiburg RNA tools: a central online resource for RNA-focused research and teaching
Nucleic Acids Research, 46(W1), W25-W29, 2018.
Results are computed with SECISDesign version 1.0 (2009-10-13) using Turner99 energies
Overview
The following parameters are used to control the execution of SECISDesign
Furthermore, additional information is available
Original Protein Sequence
 Protein sequence
Protein sequence
Your sequence of amino acids of the protein in single letter code
in which you wish to insert the selenocysteine. Indicate a stop by '#'.
The parameter constraints are: Only the IUPAC alphabet ACDEFGHIKLMNPQRSTVWY# (all capital) is allowed for specification. String length has to be in range (5,300). Maximally 1 line is allowed.
Defaults to ()
SECIS Design Constraints
 Position of Selenocystein in Protein
Position of Selenocystein in Protein
Here, you can choose the position within your amino acid sequence where you wish to insert the selenocysteine.
The parameter constraints are: Input value has to be parsable as Integer. The value must be greater than or equal to 1 and must be smaller than or equal to 300. Has to be within protein length range.
Defaults to ()
 Amino Acids to Conserve
Amino Acids to Conserve
Please insert the number and the amino acid(s) of the conserved position(s) of your given amino acid sequence. Indicate a stop by #.
Examples:
|
97 F
| means, that the F is conserved at the 97th position
|
|
| 98 S T
| means, that the 98th position is conserved to S or T
|
The parameter constraints are: Has to be in the format 'POSITION AA1 AA2 ...' per line.
Defaults to ()
 SECIS-Element
SECIS-Element
 Custom structure
Custom structure
If you have not chosen one of the six given SECIS-elements, you can create
your own one. To this end, you have to give the structure in bracket notation and
the conserved nucleotides. An example for
FdhF-std (optional)
is given below.
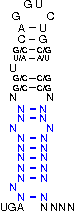
[,[[[[[{[[/((.((((....))))))\]]}]]]]]]....
NNNNNNNNNNNSSUWSCAGGUCUGSWSSNNNNNNNNNNNNNN
The following symbols can be used to define the custom SECIS element.
|
( ) |
represents a fixed bond |
|
|
G U |
represends a fixed bond for which G-U and U-G pairs are allowed
(Please note, that "G" always represents the opening bracket
and "U" represents the closing one.)
|
|
|
[ ] |
represends an optional bond (a bond that is not fixed, but
of advantage if it forms, nevertheless it is not
necessary to ensure the function of the SECIS-element)
|
|
|
. |
represents a fixed unbound position |
|
|
/ \ |
represents fixed unbound positions in an interior loop |
|
|
{ } |
represents optionally unbound positions (of advantage if
they do not bind) |
|
|
, |
represents optionally unbound positions in a bulge loop |
Note: Unbound positions in interior loops (/ \) and
optionally unbound positions in bulge loops (,) must be given by special
characters.
The parameter constraints are: String length has to be in range (0,300). Maximally 1 line is allowed. Has to be of the alphabet '.,()[]{}GU/\' and its length has to be a multiple of 3 to encode codons.
Defaults to ()
 Custom sequence
Custom sequence
Sequence constraint using
IUPAC ambiguity codes for nucleotides {ACGTURYMKWSBDHVN} with wild-card "N".
A detailed list of the codes is given below.
| IUPAC nucleotide code |
Base |
| A | Adenine |
| C | Cytosine |
| G | Guanine |
| U | Uracil |
| R | A or G |
| Y | C or U |
| S | G or C |
| W | A or U |
| K | G or U |
| M | A or C |
| B | C or G or U |
| D | A or G or U |
| H | A or C or U |
| V | A or C or G |
| N | any base |
The parameter constraints are: Only the IUPAC alphabet 'ACGURYMKWSBDHVN' is allowed for specification. If provided, it has to have the same length as the structure constraint.
Defaults to ()
Similarity Scoring
 Similarity
Similarity
Chose one of the available amino acid similarity matricies to be
used for substitution scoring.
 Insertion Penalty
Insertion Penalty
During the structure optimization, we allow insertions and deletions in the
amino acid sequence. This is to avoid contradictions between fixed positions
on the amino acid and the nucleotide level. Nevertheless, these insertions and
deletions have to be penalized. The values of these penalties are given by the
Insertion and Deletion Penalty and related to the similarity scores of PAM 250
and BLOSUM 62.
 Deletion Penalty
Deletion Penalty
During the structure optimization, we allow insertions and deletions in the
amino acid sequence. This is to avoid contradictions between fixed positions
on the amino acid and the nucleotide level. Nevertheless, these insertions and
deletions have to be penalized. The values of these penalties are given by the
Insertion and Deletion Penalty and related to the similarity scores of PAM 250
and BLOSUM 62.
RNAinverse (local search)
 Search Strategy
Search Strategy
The postprocessing is done by a local search method as implemented in RNAinverse.
During each of the following strategies, single bases or base pairs are mutated.
|
Adaptive Walk:
| During this strategy, a mutation is accepted if it results in a better
value of the objective function (e.g. folding probability).
Therefore, the adaptive walk is also called fast local search. The search terminates if no
mutation can be found which betters the objective function.
|
|
| Full Local Search:
| This approach is similar to the adaptive walk. But during the full local search,
a mutation will just be accepted if it results in a better value of the objective function AND
no other mutation exists that yields a better value. The search terminates if no
mutation can be found which betters the objective function.
|
|
| Stochastic Local Search:
| The strategy of stochastic local search has a lot in common with the adaptive walk.
Whereas the latter often gets stuck in local optima (sequences for which no mutation with a
better value of the objective function exists), the stochastic local search is allowed to mutate
to worse sequences with a fixed probability p to overcome local optima. A mutation is retained if it
results in a better value of the objective function or even if the value is worse with probability
p. We set p to 0.1. The search terminates after a fixed number of mutations. We set this number
to 500.
|
 Objective Function
Objective Function
During the postprocessing (local search), a second objective function is needed (in addition to the similarity)
to increase the folding probability of the mRNA sequence.
One of the following functions or combinations of them can be chosen:
|
mfe:
| Minimizing the distance of the minimum-free-energy-structure of the
designed sequence and the wanted structure.
|
|
| nc:
| Minimizing the average number of incorrect paired nucleotides.
|
|
| pf:
| Maximizing the probability of the designed sequence folding into the wanted
structure.
|
 Valid Similarity Fraction
Valid Similarity Fraction
During the postprocessing (local search), a new objective function is considered.
Nevertheless the similarity has to be kept clearly in mind.
Therefore you can choose the fraction of the similarity to compare with, which
must be kept during the local search (while optimizing the second objective function).
e.g.: Valid Similarity Fraction = 0.9 assures that, during local search, the new similarity is
not allowed to be lower than 90% of the compared similarity.
 Compared Similarity
Compared Similarity
During the postprocessing (local search), a new objective function is considered.
Nevertheless the similarity has to be kept clearly in mind.
Therefore you can choose the similarity which has to be compared to the values arising
during the local search.
Either you choose the start similarity (the best possible one, which arises after the
first part of the algorithm), or you decide to compare your current value with the
previous one.
 Probabilities of Bases
Probabilities of Bases
During the postprocessing (local search), single bases or base pairs are mutated.
Here, you can choose whether
- all bases should have the
same probability or
- A and U should have a higher
probability on unpaired positionen
while
C and G are more probable on
paired ones
Output Description
Here, a typical use case of SECISDesign is given: If you wish to express an eukaryotic selenoprotein in
E.coli, this is not directly possible, since there are differences between the mechanisms for inserting
selenocysteine in eukaryotic and bacterial proteins. In eukaryotes, the SECIS-element is located in the 3' UTR of the
mRNA with a distance from the UGA-codon that varies from 500 to 5300 nucleotides. In bacteria, the situation
is quite different. The SECIS-element is located immediately
downstream the UGA-codon, which implies that the SECIS-element is in the coding part of the protein.
Thus, we have the following implications. First, an eukaroytic selenoprotein cannot directly be expressed
in the
E.coli system, since it requires the design of
an appropriate SECIS-element directly after the UGA-position. Second, this design always changes the protein
sequence. Therefore, one has to make a compromise between changes in the protein sequence and the efficiency of
selenocysteine insertion (i.e. the quality of the SECIS-element).
SECISDesign searchs for similar proteins under sequential and structural constraints imposed on the mRNA by the
SECIS-elements.
Let's choose the
mammalian methionine sulfoxide reductase B (MsrB). If we wish to express it in
E.coli,
we have to change the coding mRNA such that it can form a SECIS-element and codes for a highly similar amino acid
sequence.
Input (see example):
First, the sequence of amino acids of the protein,
in which the selenocysteine should be inserted, is put into the
Amino Acid Sequence field, e.g.:
MSFCSFFGGEVFQNHFEPGVYVCAKCSYELFSSHSKYAHSSPWPAFTETIHPDSVTKC
PEKNRPEALKVSCGKCGNGLGHEFLNDGPKRGQSRFCIFSSSLKFVPKGKEAAASQGH
| Second, you can choose |
- the Position of Selenocysteine: the position (within your given amino acid sequence) on which you wish to insert
the selenoysteine (e.g. 95)
- the SECIS-Element you wish to insert (e.g.:
FdhF-std+optional)
- and optionally some restrictions about positions of your sequence which must not be
changed (e.g.: 98 S T, which means, that the 98th position is conserved to S or
T). You have to put these information into the field of the Amino Acid Conditions.
|
Third, the
Similarity measurement, e.g.:
BLOSUM62, and the values for penalizing insertions
and deletions can be chosen.
Finally, you can set some parameters, which will be used during the preprocessing step of SECISDesign.
Results:
mRNA Sequence with Structure and its Probability for the SECIS-Element region after UGA stop codon
| Wanted Structure: |
[,[[[[[{[[/((.((((....))))))\]]}]]]]]].... |
Prob.: |
| mRNA-Sequence without optimizing the stability of the structure: |
AUUUUCUCUUCGCUACCAGGUCUGGUGCCAAAAGGAAAAGAA
..(((((.((.((.((((....)))))).)).))))).....
.((((((.((.((.((((....)))))).)).)))))).... |
(0.04)
(0.19) |
| mRNA-Sequence after optimizing the stability of the structure: |
AUCUUCUCGUCGCUACCAGGUCUGGUGCCACAAGGAGCCGAA
..(((((.((.((.((((....)))))).)).)))))..... |
(0.75) |
The "mRNA-Sequence without optimizing the stability of the structure"
is the best sequence after the first step of SECISDesign. The structure below is the
wanted structure with the folding probability. If this is not the structure of minimum free energy (mfe-structure),
this one is given as well. If the mfe-structure is given in green, it is valid as well. But if
it is given in red, the mfe-structure is not valid concerning the wanted structure. The user
might decide whether this structure of minimum free energy fits his requirements anyway. The folding probability is also given.
The "mRNA-Sequence after optimizing the stability of the structure" is the gained mRNA-sequence after the
second step of SECISDesign. The structure and folding probability are given as well. Analogous to the sequence of the first step,
the structure of minimum free energy might be given. The color helps the user again to identify whether this structure is valid
or not.
Amino Acid Sequence (after SECIS insertion position)
| Original Sequence (starting at pos. 96): |
I F S S S L K F V P K G K E |
| Without optimizing the stability of the mRNA-structure: |
I F S S L P G L V P K G K E
|
| After optimizing the stability of the mRNA-structure: |
I F S S L P G L V P Q G A E
|
The "Original Sequence (starting at pos. 96)" is the considered part of
your given amino acid sequence.
The "Amino Acid Sequence without optimizing the stability of the mRNA-structure" is the resulting amino
acid sequence after the first part of SECISDesign. Changed positions are given in blue.
The "Amino Acid Sequence after optimizing the stability of the mRNA-structure" is the final amino acid
sequence encoded by an mRNA which has a higher probability to fold into the desired structure than the mRNA of the "Amino Acid
Sequence without optimizing the stability of the mRNA-structure". Changed position are given in blue as well.
Input Examples
 Custom SECIS in MsrB
Custom SECIS in MsrB
Insert a custom SECIS (which is FdhF-std (optional)) to mammalian methionine sulfoxide reductase B (MsrB). See help page for an explanation of the output.
The
example's result can be directly accessed
here Insert SECIS in MsrB
Insert SECIS in MsrB
Insert SECIS to mammalian methionine sulfoxide reductase B (MsrB). See help page for an explanation of the output.
The
example's result can be directly accessed
hereList of Changes
- 4.0.0 : SECISDesign webserver now part of Freiburg RNA tools server