Use the following button if you want to resubmit the job with
altered input or parameterization:
Input and runtime details for job 1310125 (precomputed example)
Original Protein Sequence
 Protein sequence Protein sequence | MSFCSFFGGEVFQNHFEPGVYVCAKCSYELFSSHSKYAHSSPWPAFTETIHPDSVTKCPEKNRPEALKVSCGKCGNGLGHEFLNDGPKRGQSRFCIFSSSLKFVPKGKEAAASQGH |
SECIS Design Constraints
 Position of Selenocystein in Protein Position of Selenocystein in Protein | 95 | ||
 Amino Acids to Conserve Amino Acids to Conserve | 97 F 98 S T | ||
 SECIS-Element SECIS-Element | FdhF-std (optional) | ||
 Custom structure Custom structure | not provided | ||
 Custom sequence Custom sequence | not provided |
Similarity Scoring
 Similarity Similarity | BLOSUM62 | ||
 Insertion Penalty Insertion Penalty | -10 | ||
 Deletion Penalty Deletion Penalty | -5 |
RNAinverse (local search)
 Search Strategy Search Strategy | Full Local Search | ||
 Objective Function Objective Function | mfe -> pf | ||
 Valid Similarity Fraction Valid Similarity Fraction | 0.7 | ||
 Compared Similarity Compared Similarity | Start Similarity | ||
 Probabilities of Bases Probabilities of Bases | all equal |
Job ID 1310125 (server version 4.0.0)
 | Job Submitted & Queued | @ Thu Jun 18 18:25:24 CEST 2015 | |
 | SECISDesign Started | @ Thu Jun 18 18:25:37 CEST 2015 | |
 | SECISDesign Finished & Post-Processing | @ Thu Jun 18 18:26:05 CEST 2015 | |
 | Job Completed | @ Thu Jun 18 18:26:06 CEST 2015 |
DIRECT ACCESS: https://rna.informatik.uni-freiburg.de/RetrieveResults.jsp?jobID=1310125&toolName=SECISDesign ( 30 days expiry )
Description of the job
Insert SECIS in MsrB
Insert SECIS to mammalian methionine sulfoxide reductase B (MsrB). See help page for an explanation of the output.
Output
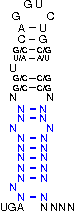
mRNA Sequence with Structure and its Probability for the SECIS-Element region after UGA stop codon
| Wanted Structure: | [,[[[[[{[[/((.((((....))))))\]]}]]]]]]....
| Prob.: |
| mRNA-Sequence without optimizing the stability of the structure: | AUUUUCUCUUCGCUACCAGGUCUGGUGCCAAAAGGAAAAGAA ..(((((.((.((.((((....)))))).)).)))))..... | (0.04) (0.19) |
| mRNA-Sequence after optimizing the stability of the structure: | AAGUUCUCUUCGCUAGCAGGUCUGCUGCCAAAAGGACAAGAA | (0.58) |
Amino Acid Sequence after selenocystein insertion position
| Original Sequence (starting at pos. 96): | I F S S S L K F V P K G K E |
| Without optimizing the stability of the mRNA-structure: |
I F S S L P G L V P K G K E
|
| After optimizing the stability of the mRNA-structure: |
K F S S L A G L L P K G Q E
|
Job resubmission
 usability assessment
usability assessment
When using SECISDesign please cite :
- Anke Busch, Sebastian Will, and Rolf Backofen
SECISDesign - A Server to Design SECIS-Elements within the Coding Sequence
Bioinformatics, 2005, 21(15), 3312-3. - Martin Raden, Syed M Ali, Omer S Alkhnbashi, Anke Busch, Fabrizio Costa, Jason A Davis, Florian Eggenhofer, Rick Gelhausen, Jens Georg, Steffen Heyne, Michael Hiller, Kousik Kundu, Robert Kleinkauf, Steffen C Lott, Mostafa M Mohamed, Alexander Mattheis, Milad Miladi, Andreas S Richter, Sebastian Will, Joachim Wolff, Patrick R Wright, and Rolf Backofen
Freiburg RNA tools: a central online resource for RNA-focused research and teaching
Nucleic Acids Research, 46(W1), W25-W29, 2018.
Results are computed with SECISDesign version 1.0 (2009-10-13) using Turner99 energies



