Class assignments -- orientation not predicted
Class assignments -- orientation not predicted
| ID | Input | Structures | Sequences | Superclass |
|---|---|---|---|---|
| 1 | UserId=Repeat1 GCATGCTGCGCGTGCGCGCGGGACTGCC |
- | - | B |
| 2 | UserId=Repeat2 GTCGCCCTCCTCGCGGAGGGCGTGGATAGAAA |
- | - | C |
| 3 | UserId=Repeat3 GAATCTCAAAGAGAGGATTGAAAC |
- | - | D |
| 4 | UserId=Repeat4 GTCTCCTTTACCACTTCCCCGAGAGGGGATTGAAAG |
motif 4 | - | E |
| 5 | UserId=Repeat5 TTTCCGATTTTTCGGCTCGCAGAAGAGCACTGAAA |
motif 17 | - | E |
| 6 | UserId=Repeat6 GATGCAACTAAGAAAGAATTGAAGC |
- | - | D |
| 7 | UserId=Repeat7 CRISPRmap=Tequ_A_G_3_X_F6 TTAACTACCGTATAGGTAGTTTAGA |
- | family 6 | - |
| 8 | UserId=Repeat8 GTCTCAACGGCCTAAGCGGCCAGCGGGTCTGAAA |
- | - | C |
 Close up on structure motifs
Close up on structure motifs
Details on motif 1:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 2:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 3:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
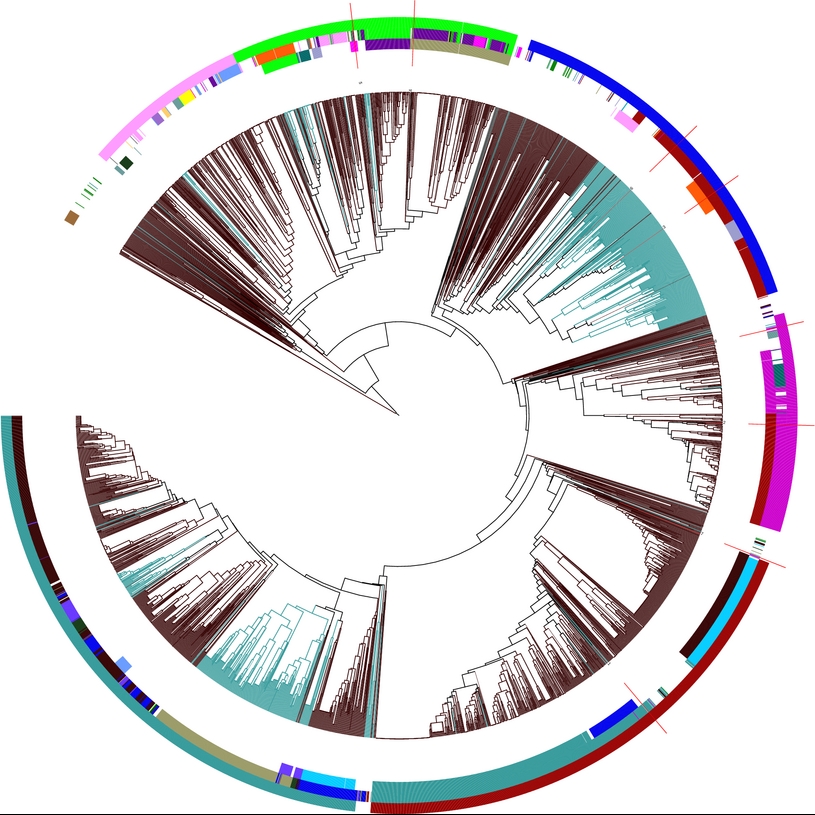
Details on motif 4:
|
Clustering tree:
|
Comments:
| ||||||||||||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs Your sequences are highlighted in red! |
Alignment:
|
Consensus MFE structure:
|
Update: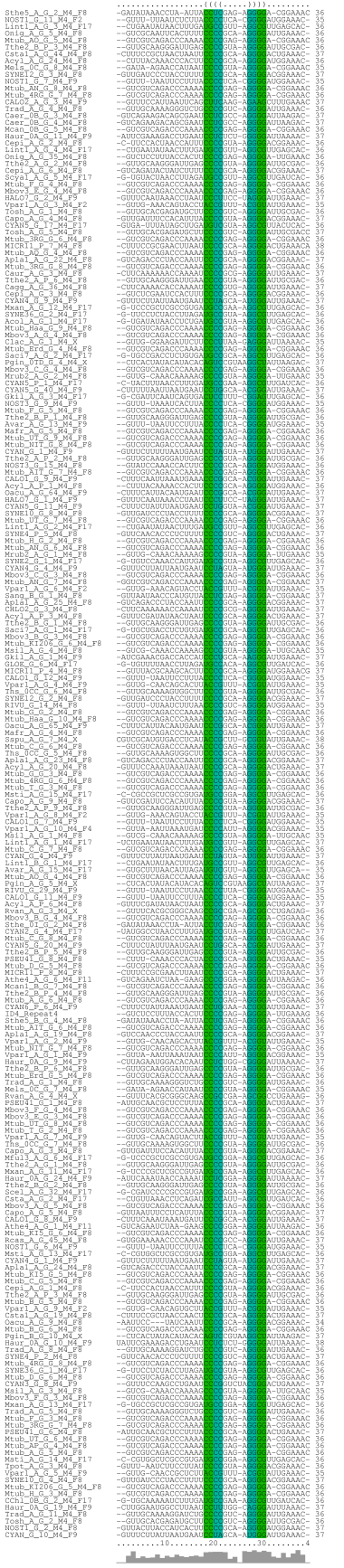
|
Update: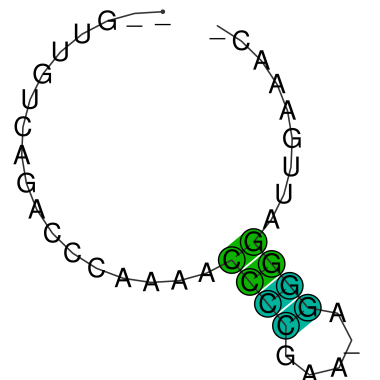
|
Details on motif 5:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 6:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 7:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 8:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 9:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 10:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 11:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 12:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 13:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 14:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 15:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
Details on motif 16:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
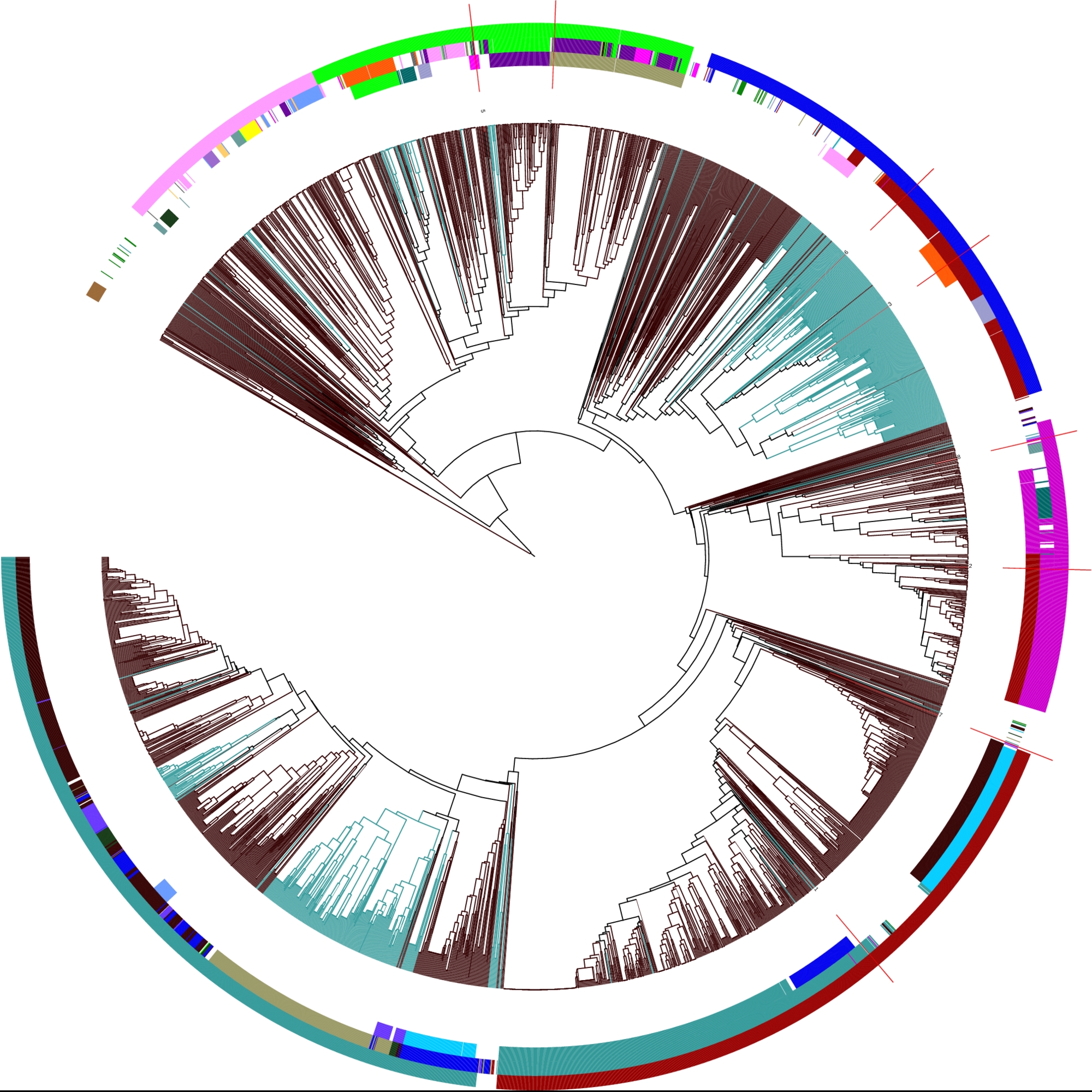
Details on motif 17:
|
Clustering tree: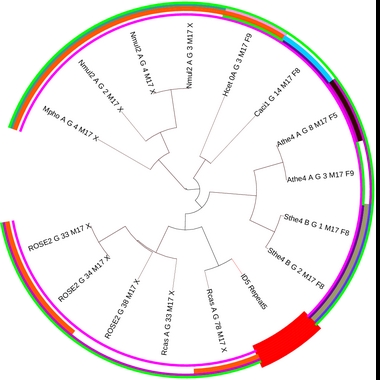
|
Comments:
| ||||||||||||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs Your sequences are highlighted in red! |
Alignment:
|
Consensus MFE structure:
|
Update: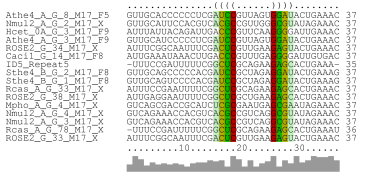
|
Update: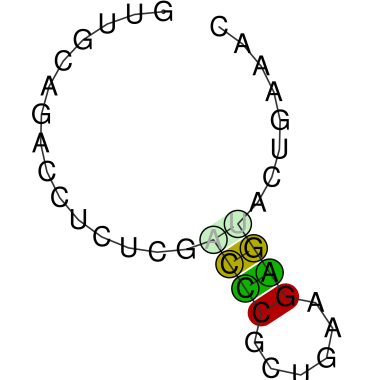
|
Details on motif 18:
|
Clustering tree: |
Comments:
| |||||||||||||||||
RNAz alignment statistics:
|
Tree-Legend:
(from inside to outside) Structure motifs | ||||||||||||||||||
Alignment:
|
Consensus MFE structure:
|
 Close up on sequence families
Close up on sequence families
Details on family 1:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 2:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 3:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 4:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 5:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 6:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 7:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 8:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 9:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 10:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 11:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 12:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 13:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 14:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 15:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 16:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 17:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 18:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 19:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 20:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 21:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
Details on family 22:
|
Comments:
| |||||||||||||||
Sequence logo of all aligned family members:
|
||||||||||||||||
 Downloads
Downloads
 CRISPRmap version used for prediction
CRISPRmap version used for prediction
Input and runtime details for job 0788874 (precomputed example)
Input Parameters
 Sequence Input in FASTA Format Sequence Input in FASTA Format | [.fa] | ||
 Optimize reading direction of input sequences Optimize reading direction of input sequences | no |
Job ID 0788874 (server version 3.4.2)
 | Job Submitted & Queued | @ Mon Dec 15 12:36:20 CET 2014 | |
 | CRISPRmap Started | @ Mon Dec 15 12:36:31 CET 2014 | |
 | CRISPRmap Finished & Post-Processing | @ Mon Dec 15 13:02:26 CET 2014 | |
 | Job Completed | @ Mon Dec 15 13:02:36 CET 2014 |
When using CRISPRmap please cite :
- Omer S. Alkhnbashi, Fabrizio Costa, Shiraz A. Shah, Roger A. Garrett, Sita J. Saunders, and Rolf Backofen.
CRISPRstrand: predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci
Bioinformatics, 2014, 30(17), 489-496. - Sita J. Lange, Omer S. Alkhnbashi, Dominic Rose, Sebastian Will and Rolf Backofen.
CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems
Nucleic Acids Research, 2013, 41(17), 8034-8044. - Martin Raden, Syed M Ali, Omer S Alkhnbashi, Anke Busch, Fabrizio Costa, Jason A Davis, Florian Eggenhofer, Rick Gelhausen, Jens Georg, Steffen Heyne, Michael Hiller, Kousik Kundu, Robert Kleinkauf, Steffen C Lott, Mostafa M Mohamed, Alexander Mattheis, Milad Miladi, Andreas S Richter, Sebastian Will, Joachim Wolff, Patrick R Wright, and Rolf Backofen
Freiburg RNA tools: a central online resource for RNA-focused research and teaching
Nucleic Acids Research, 46(W1), W25-W29, 2018.